Search
Supporting Material for  BioQuery Tool Discovery System (Bio-TDS) The Bio-TDS (BioQuery Tools Discovery Systems) has been developed to assist researchers in retrieving the most applicable analytic tools by allowing them to formulate their questions as free text. The Bio-TDS is a flexible retrieval system that affords users from multiple bioscience domains ( e.g. genomic, proteomic, bio-imaging) the ability to query over 12,000 analytic tool descriptions integrated from well-established, community repositories. One of the primary components of the Bio-TDS system is the ontology and natural language processing workflow for annotation, curation, query processing, and evaluation. The Bio-TDS’s scientific impact was evaluated using sample questions posed by researchers retrieved from Biostars, a site focusing on biological data analysis. The Bio-TDS was compared to five similar bioscience analytic tool retrieval systems with the Bio-TDS outperforming the others in terms of relevance and completeness. The Bio-TDS offers researcher the capacity to associate their bioscience question with the most relevant computational toolsets required for the data analysis in their knowledge discovery process.
BioQuery Tool Discovery System (Bio-TDS) The Bio-TDS (BioQuery Tools Discovery Systems) has been developed to assist researchers in retrieving the most applicable analytic tools by allowing them to formulate their questions as free text. The Bio-TDS is a flexible retrieval system that affords users from multiple bioscience domains ( e.g. genomic, proteomic, bio-imaging) the ability to query over 12,000 analytic tool descriptions integrated from well-established, community repositories. One of the primary components of the Bio-TDS system is the ontology and natural language processing workflow for annotation, curation, query processing, and evaluation. The Bio-TDS’s scientific impact was evaluated using sample questions posed by researchers retrieved from Biostars, a site focusing on biological data analysis. The Bio-TDS was compared to five similar bioscience analytic tool retrieval systems with the Bio-TDS outperforming the others in terms of relevance and completeness. The Bio-TDS offers researcher the capacity to associate their bioscience question with the most relevant computational toolsets required for the data analysis in their knowledge discovery process. 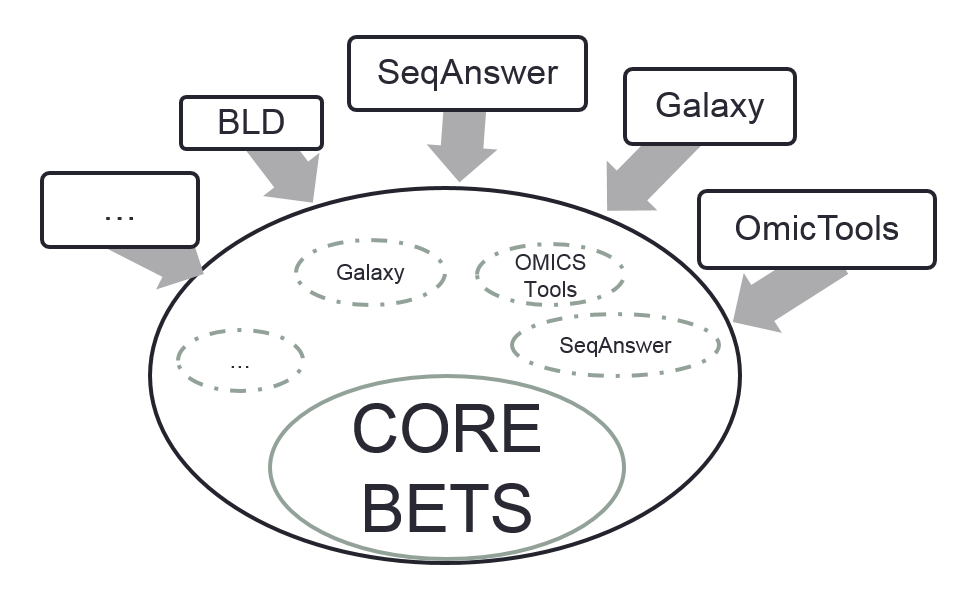 Figure S1a: BETS design Overview Bioinformatics Elaborated Tools Specifications (BETS) provides a standard for analytic tool descriptions. The analytic tool descriptions (i.e. metadata) gathered from community tool repositories integrated into the Bio-TDS are stored in JSON format using the BETS standard. This standard consists of core BETS attributes and domains/repositories specifics attributes (see Figure S1) The core BETS attributes are manually mapped to the repository attribute (more here
Figure S1a: BETS design Overview Bioinformatics Elaborated Tools Specifications (BETS) provides a standard for analytic tool descriptions. The analytic tool descriptions (i.e. metadata) gathered from community tool repositories integrated into the Bio-TDS are stored in JSON format using the BETS standard. This standard consists of core BETS attributes and domains/repositories specifics attributes (see Figure S1) The core BETS attributes are manually mapped to the repository attribute (more here  )
) 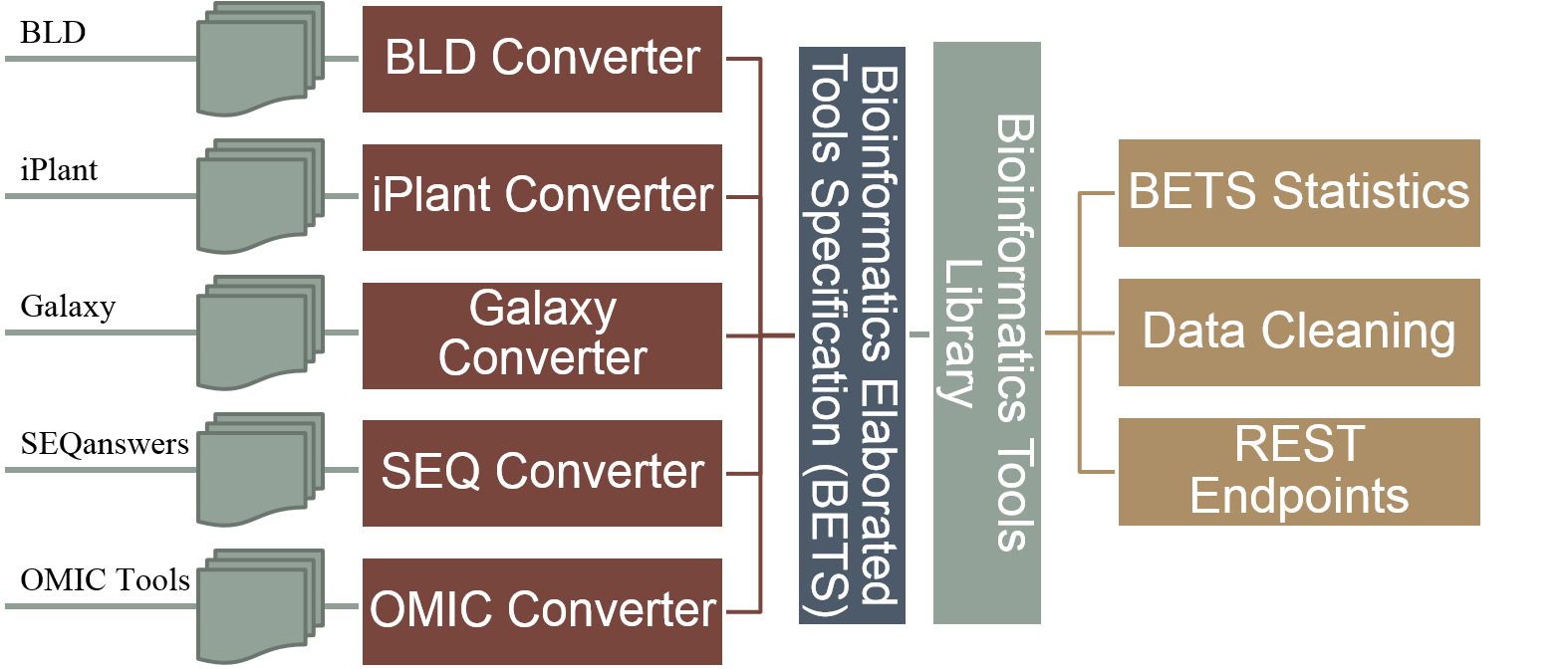 Figure S1b: BETS Converter
Figure S1b: BETS Converter 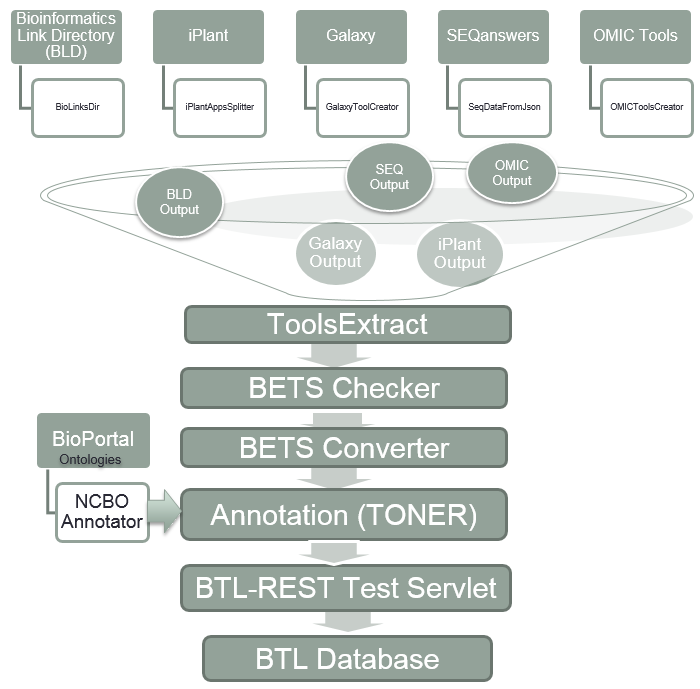 Figure S2: Bio-TDS extraction workflow A rule-based (or predicate) semi-automatic curation process has been developed for the Bio-TDS by combining human inspection and data mining methods. Rules are generated manually and are applied in a very specific order. For example, some rules such as [if tool type is ‘not tool’, then remove the tool from the repository] are applied during the extraction process. This rule, for example, results in the number of tools extracted from OMICtools to be reduced from 10,941 to 8,423. This is because OMICtools defines ‘links to a literature’ and other resource types as tools. Even if the scope of this rule is limited to just one attribute, its consistency among repositories and impact on the curation process is relevant and effective. Following this logic, several categories were identified and used to lead the development of the rules: community/domain related rules, design related rules, and human intuitive rules (Table S2). After integration and curation, we have a very good improvement in our repository content (more
Figure S2: Bio-TDS extraction workflow A rule-based (or predicate) semi-automatic curation process has been developed for the Bio-TDS by combining human inspection and data mining methods. Rules are generated manually and are applied in a very specific order. For example, some rules such as [if tool type is ‘not tool’, then remove the tool from the repository] are applied during the extraction process. This rule, for example, results in the number of tools extracted from OMICtools to be reduced from 10,941 to 8,423. This is because OMICtools defines ‘links to a literature’ and other resource types as tools. Even if the scope of this rule is limited to just one attribute, its consistency among repositories and impact on the curation process is relevant and effective. Following this logic, several categories were identified and used to lead the development of the rules: community/domain related rules, design related rules, and human intuitive rules (Table S2). After integration and curation, we have a very good improvement in our repository content (more  ) Table S2: Selected curation rules
) Table S2: Selected curation rules 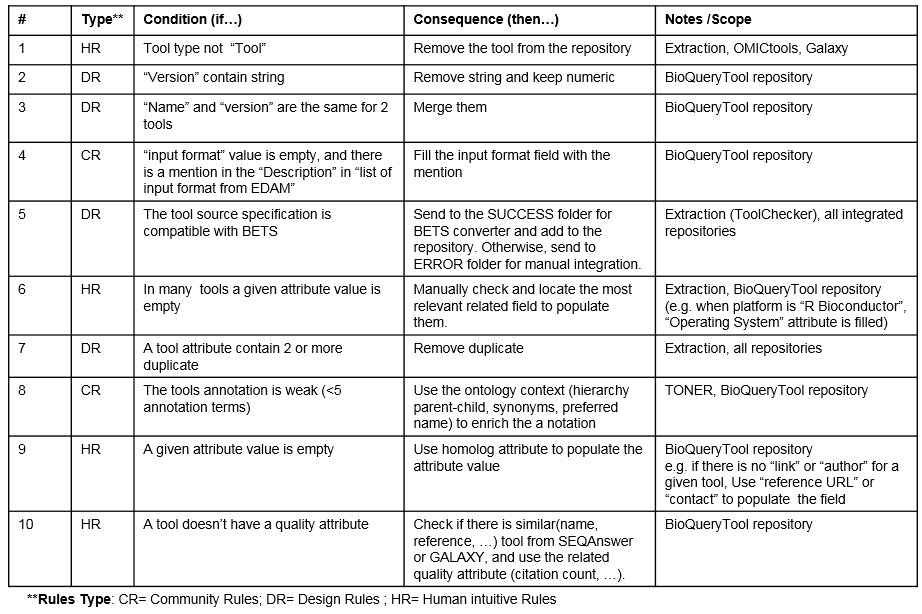 Community or domain related rules--are governed by information representation and information management in each community. For scientific communities such as bioinformatics, EDAM ontology is an important semantic and lexical information representation. These considerations led to the development of rules such as [if “input format” value is empty, and there is a mention in the “Description” in “list of input format from EDAM”, then fill the input format field with the mention]. Ontologies also allow us to check relationships between terms. Therefore, a relationship such as “synonym” from the ontology can help to populate some missing value or remove redundant data. Design based rules include constraints related to data integration issues. The key goal is to keep the integrated database lossless when applying operations such as merging duplicates. For example, considering the following rule [if “Name” and “version” are the same for two tools then, merge them]. This definition of tool similarity allows for a simple string comparison of two attributes (name, version) using simple edit distance. During and after the integration of analytic tool definitions, missing data management rules are used to minimize the empty values in the Bio-TDS repository. Human inspection remains the key and most accurate action during the curation process. This include rules for consistency checking (i.e. no contradiction, complete and close rules), data quality overview, and new intuitive rules generation (i.e. looking at data and statistics, some rules can be inferred, and human walkthrough suggestions. A simple Web UI was developed to allow contributors to add curation suggestions. At the current development stage, the Bio-TDS team has already identified 10 key rules.The application of that rule set has helped to improve the repository accuracy by removing duplicates and invalid tools (from 16,458 tools to 12,000), removing inaccurate attributes values, and filling in missing information. This has achieved an overall improvement rate of ~30%.
Community or domain related rules--are governed by information representation and information management in each community. For scientific communities such as bioinformatics, EDAM ontology is an important semantic and lexical information representation. These considerations led to the development of rules such as [if “input format” value is empty, and there is a mention in the “Description” in “list of input format from EDAM”, then fill the input format field with the mention]. Ontologies also allow us to check relationships between terms. Therefore, a relationship such as “synonym” from the ontology can help to populate some missing value or remove redundant data. Design based rules include constraints related to data integration issues. The key goal is to keep the integrated database lossless when applying operations such as merging duplicates. For example, considering the following rule [if “Name” and “version” are the same for two tools then, merge them]. This definition of tool similarity allows for a simple string comparison of two attributes (name, version) using simple edit distance. During and after the integration of analytic tool definitions, missing data management rules are used to minimize the empty values in the Bio-TDS repository. Human inspection remains the key and most accurate action during the curation process. This include rules for consistency checking (i.e. no contradiction, complete and close rules), data quality overview, and new intuitive rules generation (i.e. looking at data and statistics, some rules can be inferred, and human walkthrough suggestions. A simple Web UI was developed to allow contributors to add curation suggestions. At the current development stage, the Bio-TDS team has already identified 10 key rules.The application of that rule set has helped to improve the repository accuracy by removing duplicates and invalid tools (from 16,458 tools to 12,000), removing inaccurate attributes values, and filling in missing information. This has achieved an overall improvement rate of ~30%. 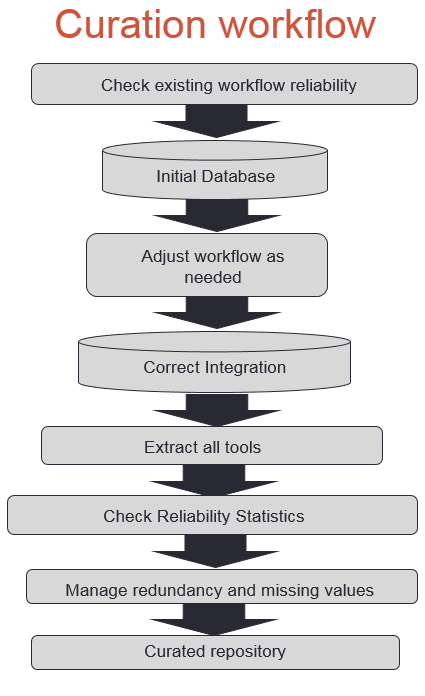
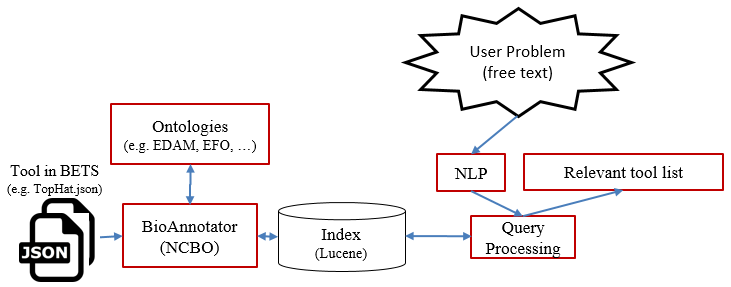
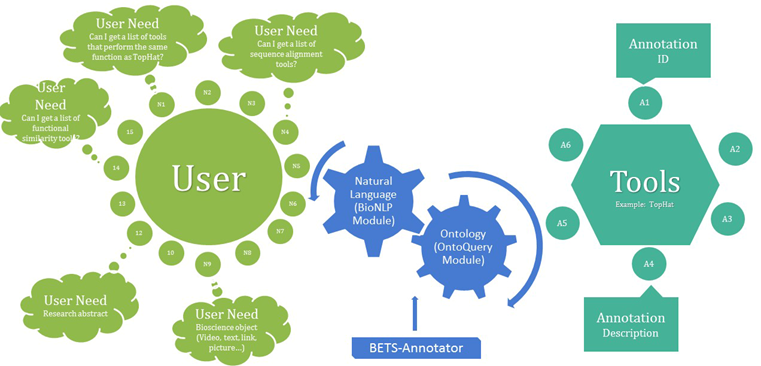 Figure S4a: Bio-TDS System Workflow
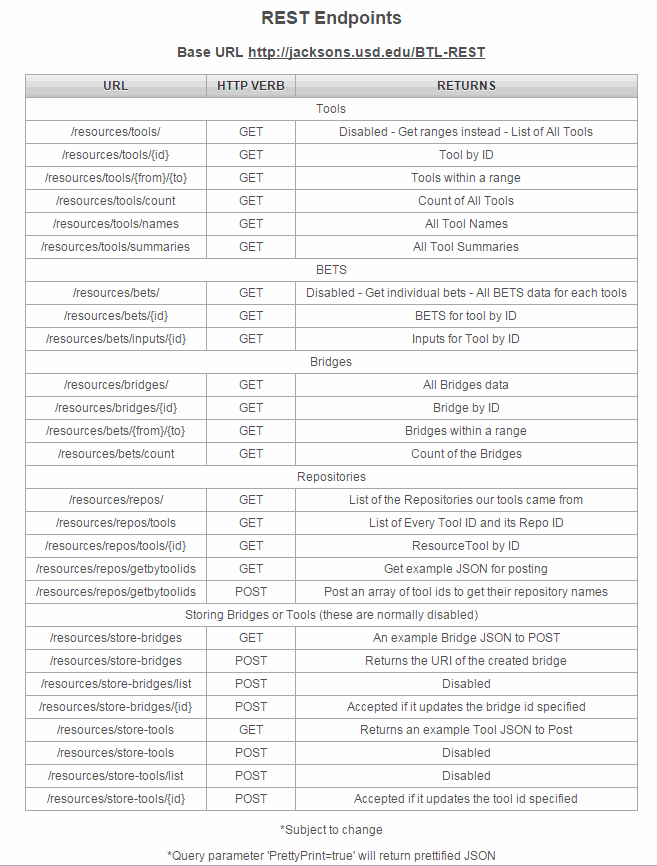
Figure S4a: Bio-TDS System Workflow 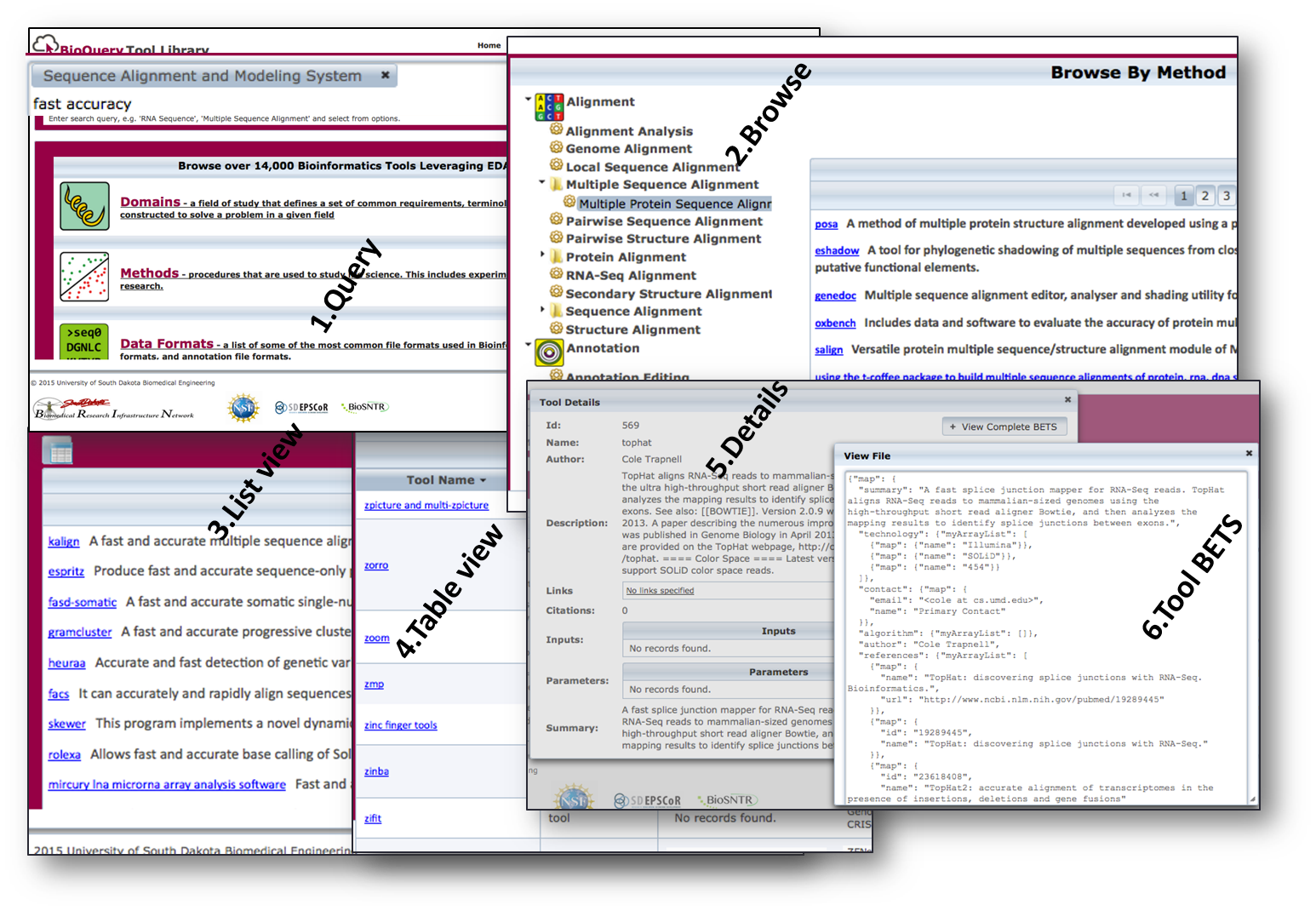 Figure S4b: Bio-TDS Display View Options Table S4: REST Endpoints
Figure S4b: Bio-TDS Display View Options Table S4: REST Endpoints  Note: The data used for the current tests have been collected from the associated repositories in December 2015. Because each repository may have been updated, the reproducibility should consider repository versions for accuracy. Figure S5a: Bio-TDS Evaluation and Comparison Overview (more here
Note: The data used for the current tests have been collected from the associated repositories in December 2015. Because each repository may have been updated, the reproducibility should consider repository versions for accuracy. Figure S5a: Bio-TDS Evaluation and Comparison Overview (more here  )
) 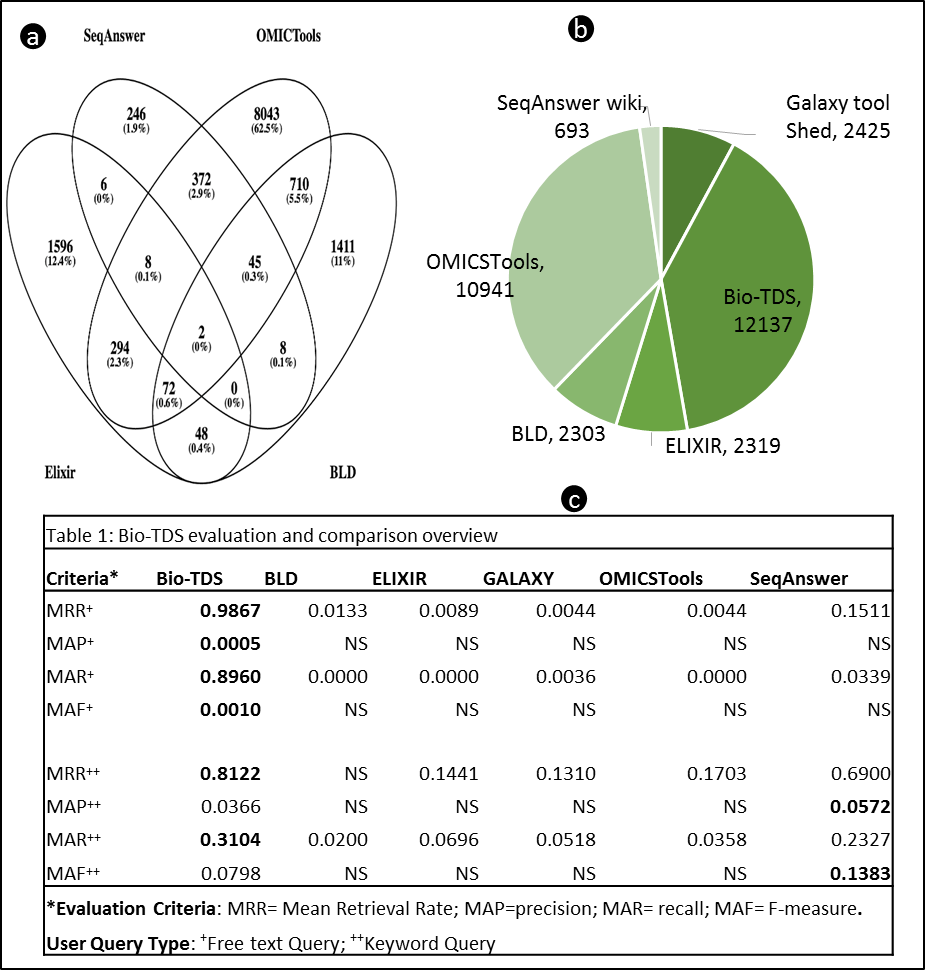 Figure S5c shows the existing repository statistic (b), as of May 2016, the tools count overlap Venn diagram (a), and repositories comparison overview (c).
Figure S5c shows the existing repository statistic (b), as of May 2016, the tools count overlap Venn diagram (a), and repositories comparison overview (c).  Note: "NS" value in a given evaluation criteria (Precision, Recall,...) indicates limited data point (missing >40% data points compare the variable dataset size) to compute an accurate meaningful criteria value. This is due to a low retrieval rate in the related repository (e.g. no result return for the query). Figure S5b: Bio-TDS Retrieval Rate Comparison
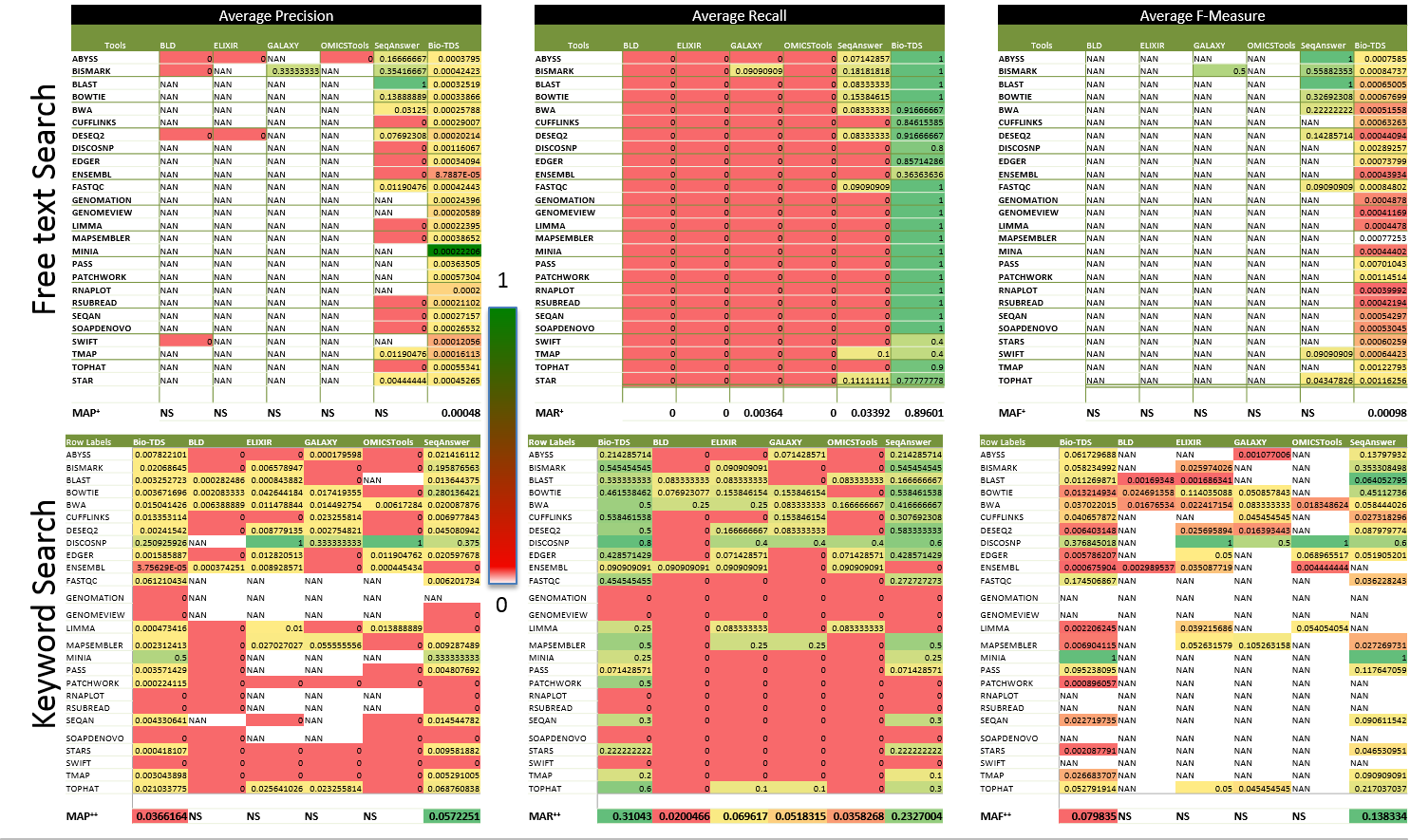
Note: "NS" value in a given evaluation criteria (Precision, Recall,...) indicates limited data point (missing >40% data points compare the variable dataset size) to compute an accurate meaningful criteria value. This is due to a low retrieval rate in the related repository (e.g. no result return for the query). Figure S5b: Bio-TDS Retrieval Rate Comparison 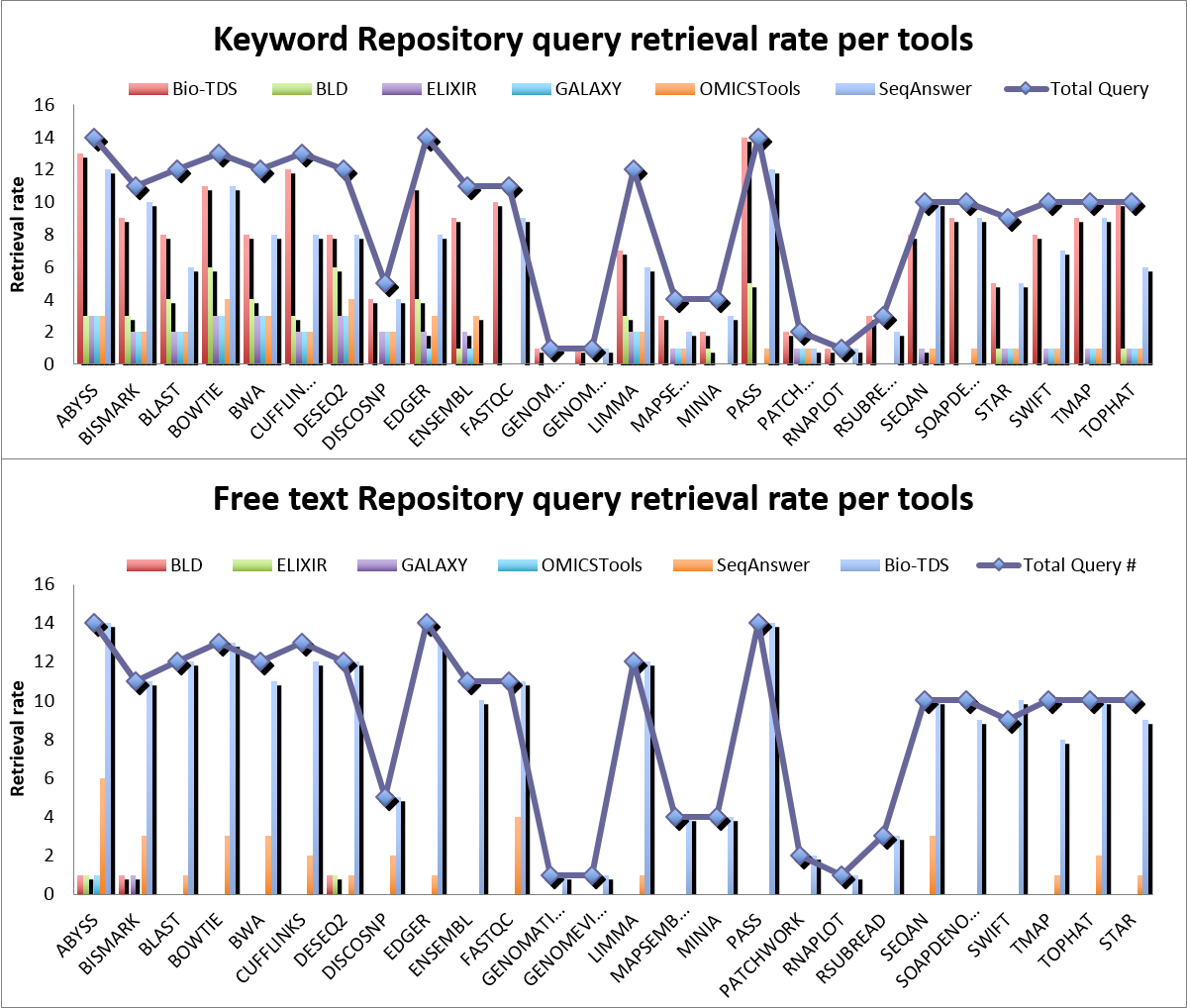 Figure S5c: Bio-TDS exactness (Precision) and completeness (Recall) Comparison (more here
Figure S5c: Bio-TDS exactness (Precision) and completeness (Recall) Comparison (more here  )
)  Back to Top
Back to Top 



 BioQuery Tool Discovery System (Bio-TDS) The Bio-TDS (BioQuery Tools Discovery Systems) has been developed to assist researchers in retrieving the most applicable analytic tools by allowing them to formulate their questions as free text. The Bio-TDS is a flexible retrieval system that affords users from multiple bioscience domains ( e.g. genomic, proteomic, bio-imaging) the ability to query over 12,000 analytic tool descriptions integrated from well-established, community repositories. One of the primary components of the Bio-TDS system is the ontology and natural language processing workflow for annotation, curation, query processing, and evaluation. The Bio-TDS’s scientific impact was evaluated using sample questions posed by researchers retrieved from Biostars, a site focusing on biological data analysis. The Bio-TDS was compared to five similar bioscience analytic tool retrieval systems with the Bio-TDS outperforming the others in terms of relevance and completeness. The Bio-TDS offers researcher the capacity to associate their bioscience question with the most relevant computational toolsets required for the data analysis in their knowledge discovery process.
BioQuery Tool Discovery System (Bio-TDS) The Bio-TDS (BioQuery Tools Discovery Systems) has been developed to assist researchers in retrieving the most applicable analytic tools by allowing them to formulate their questions as free text. The Bio-TDS is a flexible retrieval system that affords users from multiple bioscience domains ( e.g. genomic, proteomic, bio-imaging) the ability to query over 12,000 analytic tool descriptions integrated from well-established, community repositories. One of the primary components of the Bio-TDS system is the ontology and natural language processing workflow for annotation, curation, query processing, and evaluation. The Bio-TDS’s scientific impact was evaluated using sample questions posed by researchers retrieved from Biostars, a site focusing on biological data analysis. The Bio-TDS was compared to five similar bioscience analytic tool retrieval systems with the Bio-TDS outperforming the others in terms of relevance and completeness. The Bio-TDS offers researcher the capacity to associate their bioscience question with the most relevant computational toolsets required for the data analysis in their knowledge discovery process. Links to the Support Material Documentation
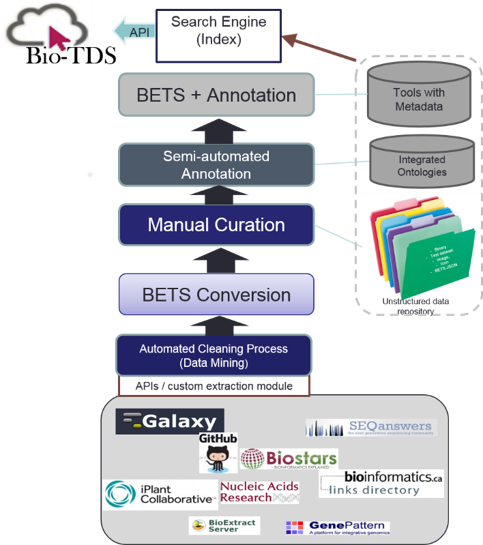
High-level view of the Bio-TDS architecture
S1 - BETS Specification description and manipulation
Back to Top Figure S1a: BETS design Overview Bioinformatics Elaborated Tools Specifications (BETS) provides a standard for analytic tool descriptions. The analytic tool descriptions (i.e. metadata) gathered from community tool repositories integrated into the Bio-TDS are stored in JSON format using the BETS standard. This standard consists of core BETS attributes and domains/repositories specifics attributes (see Figure S1) The core BETS attributes are manually mapped to the repository attribute (more here
Figure S1a: BETS design Overview Bioinformatics Elaborated Tools Specifications (BETS) provides a standard for analytic tool descriptions. The analytic tool descriptions (i.e. metadata) gathered from community tool repositories integrated into the Bio-TDS are stored in JSON format using the BETS standard. This standard consists of core BETS attributes and domains/repositories specifics attributes (see Figure S1) The core BETS attributes are manually mapped to the repository attribute (more here  )
)  Figure S1b: BETS Converter
Figure S1b: BETS Converter S2 - Resources extraction and semi-automatics curation
Back to Top The Bio-TDS combines bioinformatics tools from five other repositories and stores them in one central location, following BETS (Bioinformatics Elaborated Tool Specification). There are six main modules that convert the data from each of the five repositories into BETS tools and store the new tools into the Bio-TDS database. The BETS Checker is a Java application that tests the compatibility of a tool with the BETS specification. A tool is considered “compatible” if it is in the format specified by the specific BETS converter. For example, the system contains a mapper called Galaxy Converter. A tool from the Galaxy Tool Shed can only be “compatible” if it matches the predefined Galaxy format. Figure S2: Bio-TDS extraction workflow A rule-based (or predicate) semi-automatic curation process has been developed for the Bio-TDS by combining human inspection and data mining methods. Rules are generated manually and are applied in a very specific order. For example, some rules such as [if tool type is ‘not tool’, then remove the tool from the repository] are applied during the extraction process. This rule, for example, results in the number of tools extracted from OMICtools to be reduced from 10,941 to 8,423. This is because OMICtools defines ‘links to a literature’ and other resource types as tools. Even if the scope of this rule is limited to just one attribute, its consistency among repositories and impact on the curation process is relevant and effective. Following this logic, several categories were identified and used to lead the development of the rules: community/domain related rules, design related rules, and human intuitive rules (Table S2). After integration and curation, we have a very good improvement in our repository content (more
Figure S2: Bio-TDS extraction workflow A rule-based (or predicate) semi-automatic curation process has been developed for the Bio-TDS by combining human inspection and data mining methods. Rules are generated manually and are applied in a very specific order. For example, some rules such as [if tool type is ‘not tool’, then remove the tool from the repository] are applied during the extraction process. This rule, for example, results in the number of tools extracted from OMICtools to be reduced from 10,941 to 8,423. This is because OMICtools defines ‘links to a literature’ and other resource types as tools. Even if the scope of this rule is limited to just one attribute, its consistency among repositories and impact on the curation process is relevant and effective. Following this logic, several categories were identified and used to lead the development of the rules: community/domain related rules, design related rules, and human intuitive rules (Table S2). After integration and curation, we have a very good improvement in our repository content (more  ) Table S2: Selected curation rules
) Table S2: Selected curation rules  Community or domain related rules--are governed by information representation and information management in each community. For scientific communities such as bioinformatics, EDAM ontology is an important semantic and lexical information representation. These considerations led to the development of rules such as [if “input format” value is empty, and there is a mention in the “Description” in “list of input format from EDAM”, then fill the input format field with the mention]. Ontologies also allow us to check relationships between terms. Therefore, a relationship such as “synonym” from the ontology can help to populate some missing value or remove redundant data. Design based rules include constraints related to data integration issues. The key goal is to keep the integrated database lossless when applying operations such as merging duplicates. For example, considering the following rule [if “Name” and “version” are the same for two tools then, merge them]. This definition of tool similarity allows for a simple string comparison of two attributes (name, version) using simple edit distance. During and after the integration of analytic tool definitions, missing data management rules are used to minimize the empty values in the Bio-TDS repository. Human inspection remains the key and most accurate action during the curation process. This include rules for consistency checking (i.e. no contradiction, complete and close rules), data quality overview, and new intuitive rules generation (i.e. looking at data and statistics, some rules can be inferred, and human walkthrough suggestions. A simple Web UI was developed to allow contributors to add curation suggestions. At the current development stage, the Bio-TDS team has already identified 10 key rules.The application of that rule set has helped to improve the repository accuracy by removing duplicates and invalid tools (from 16,458 tools to 12,000), removing inaccurate attributes values, and filling in missing information. This has achieved an overall improvement rate of ~30%.
Community or domain related rules--are governed by information representation and information management in each community. For scientific communities such as bioinformatics, EDAM ontology is an important semantic and lexical information representation. These considerations led to the development of rules such as [if “input format” value is empty, and there is a mention in the “Description” in “list of input format from EDAM”, then fill the input format field with the mention]. Ontologies also allow us to check relationships between terms. Therefore, a relationship such as “synonym” from the ontology can help to populate some missing value or remove redundant data. Design based rules include constraints related to data integration issues. The key goal is to keep the integrated database lossless when applying operations such as merging duplicates. For example, considering the following rule [if “Name” and “version” are the same for two tools then, merge them]. This definition of tool similarity allows for a simple string comparison of two attributes (name, version) using simple edit distance. During and after the integration of analytic tool definitions, missing data management rules are used to minimize the empty values in the Bio-TDS repository. Human inspection remains the key and most accurate action during the curation process. This include rules for consistency checking (i.e. no contradiction, complete and close rules), data quality overview, and new intuitive rules generation (i.e. looking at data and statistics, some rules can be inferred, and human walkthrough suggestions. A simple Web UI was developed to allow contributors to add curation suggestions. At the current development stage, the Bio-TDS team has already identified 10 key rules.The application of that rule set has helped to improve the repository accuracy by removing duplicates and invalid tools (from 16,458 tools to 12,000), removing inaccurate attributes values, and filling in missing information. This has achieved an overall improvement rate of ~30%. 
S3 - TONER: Tools ontology-based annotation
Back to Top
S4- Bio-TDS Query processing workflow and programmatic access
Back to Top Figure S4a: Bio-TDS System Workflow
Figure S4a: Bio-TDS System Workflow  Figure S4b: Bio-TDS Display View Options Table S4: REST Endpoints
Figure S4b: Bio-TDS Display View Options Table S4: REST Endpoints S5 - Bio-TDS Evaluation and comparison
Back to Top Note: The data used for the current tests have been collected from the associated repositories in December 2015. Because each repository may have been updated, the reproducibility should consider repository versions for accuracy. Figure S5a: Bio-TDS Evaluation and Comparison Overview (more here
Note: The data used for the current tests have been collected from the associated repositories in December 2015. Because each repository may have been updated, the reproducibility should consider repository versions for accuracy. Figure S5a: Bio-TDS Evaluation and Comparison Overview (more here  )
)  Figure S5c shows the existing repository statistic (b), as of May 2016, the tools count overlap Venn diagram (a), and repositories comparison overview (c).
Figure S5c shows the existing repository statistic (b), as of May 2016, the tools count overlap Venn diagram (a), and repositories comparison overview (c).  Note: "NS" value in a given evaluation criteria (Precision, Recall,...) indicates limited data point (missing >40% data points compare the variable dataset size) to compute an accurate meaningful criteria value. This is due to a low retrieval rate in the related repository (e.g. no result return for the query). Figure S5b: Bio-TDS Retrieval Rate Comparison
Note: "NS" value in a given evaluation criteria (Precision, Recall,...) indicates limited data point (missing >40% data points compare the variable dataset size) to compute an accurate meaningful criteria value. This is due to a low retrieval rate in the related repository (e.g. no result return for the query). Figure S5b: Bio-TDS Retrieval Rate Comparison  Figure S5c: Bio-TDS exactness (Precision) and completeness (Recall) Comparison (more here
Figure S5c: Bio-TDS exactness (Precision) and completeness (Recall) Comparison (more here  )
)  Back to Top
Back to Top Acknowledgements




© 2016 University of South Dakota Biomedical Engineering