Search
 BioQuery Tool Discovery System (Bio-TDS) Back to Top
BioQuery Tool Discovery System (Bio-TDS) Back to Top Search by Keyword
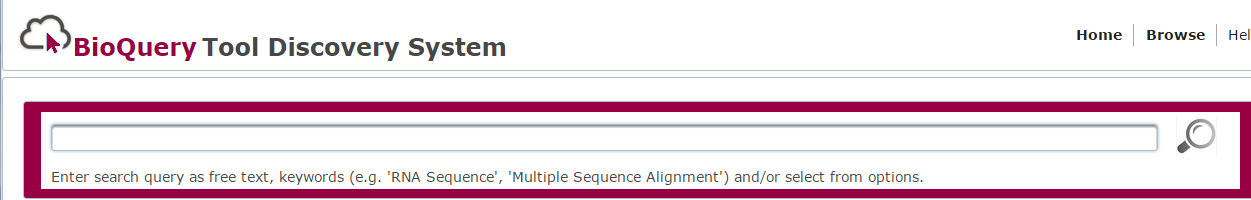
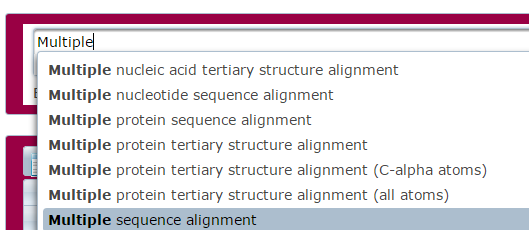
- Enter you search term(s) into the text box. The system will allow you to select from a predefined list of keywords.
- The system will provide you with a list of ontology terms.
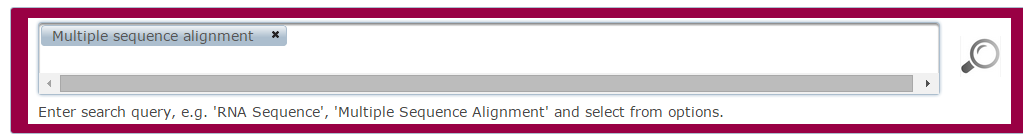
- Select search term(s) from ontology list.
- Click the
 search button.
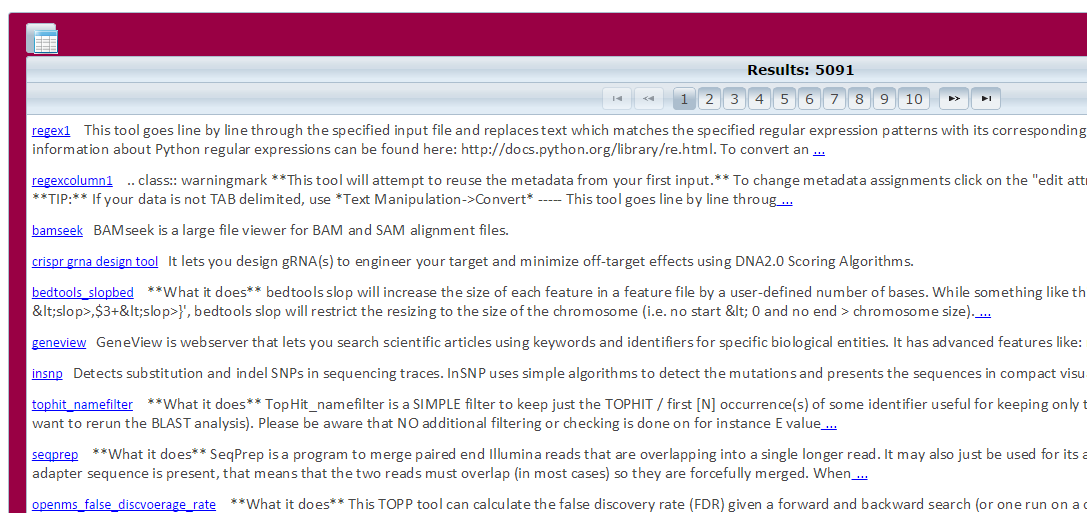
search button. - System will display a list of results.
- Click on the tool name in the results list for more detail about the tool.
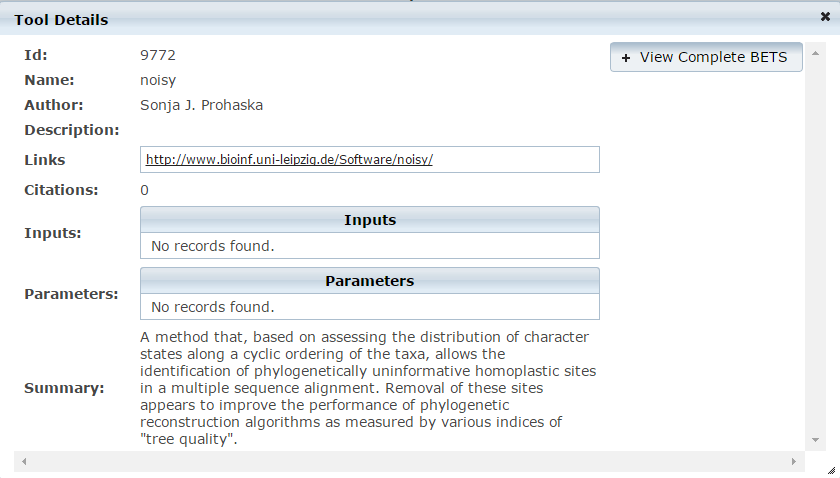
Search Using Free Text
- Enter free text into the search text box. For example, "What aligner should I use for mapping bacterial RNA-Seq data?"
- Click the
 search button.
search button. - System will display a list of results.
- Click on the tool name in results list for more detail about the tool.
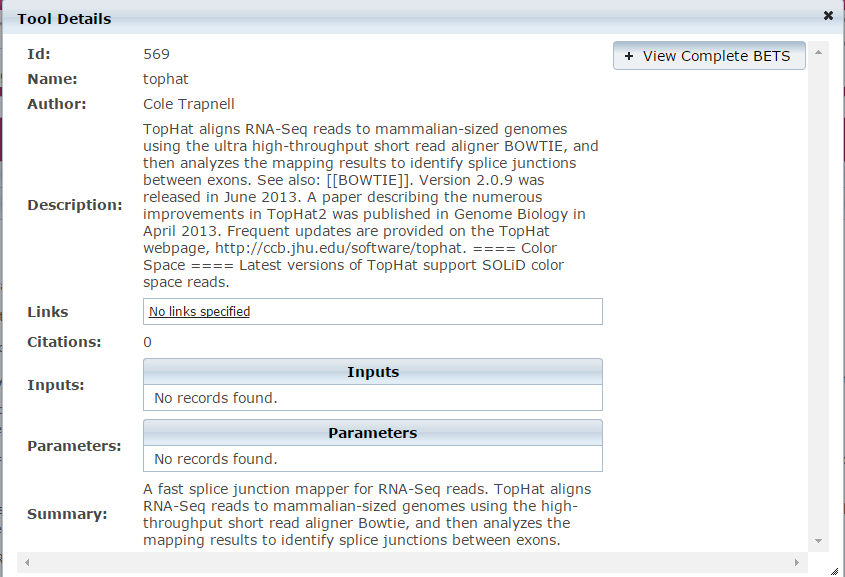
- Click on
 button to see the underling BETS tool specification.
button to see the underling BETS tool specification.

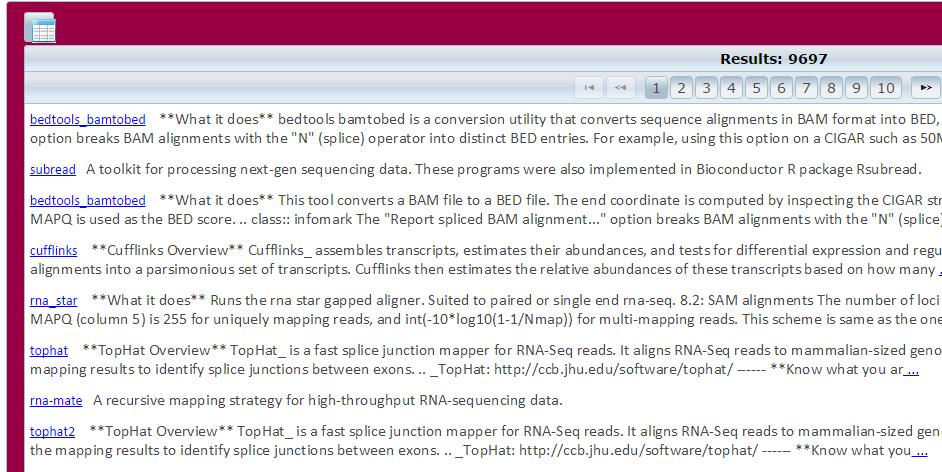
Browse by Domains, Methods, or Data Formats
- Click the Browse option located on the upper right corner of the screen.
- Click on Domains
- Expand the Biology node in the browse tree on the left.
- Select the Molecular Biology node.



Bio-TDS Overview
The aim of our BioQuery Tool project is to address some of the Bioinformatics educational challenges by creating a comprehensive, annotated Bioinformatics analytic query tool discovery system.
BioQuery Tool Discovery System (Bio-TDS) Overview
The BioQuery Tool Discovery System (Bio-TDS) is comprised of a list of tool descriptions gathered from Galaxy , iPlant Collaborative , Bioinformatics Link Directory which contains the Nucleic Acid Research Tool List, SEQAnswers, and OMIC Tools by leveraging their respective API functionality.
An ontology annotation system has been added to the BLT to enable robust searching capabilities. The ontologies to be integrated into the annotation system are: EDAM , SWO, and NGSOnto. EDAM is an ontology of concepts that are prevalent within bioinformatics, including types of data, data identifiers, data formats, operations and topics (http://edamontology.org/). The Software Ontology (SWO) is a resource for describing analytic tools, their types, tasks, versions, data requirements and provenance (http://theswo.sourceforge.net/). NGSOnto is an ontology to describe workflows from DNA extraction to contigs in a Whole Genome Sequence experiment (http://darwin.phyloviz.net/~msilva/NGSonto/).
Back to TopAcknowledgements




© 2016 University of South Dakota Biomedical Engineering
